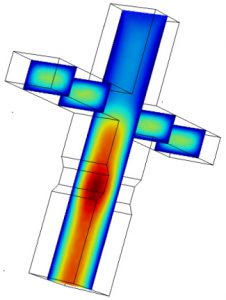
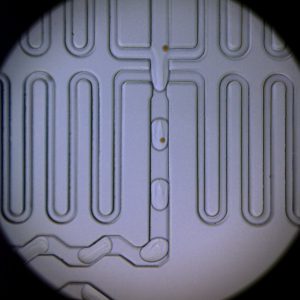
Microfluidic devices used here were all designed using AutoCAD software, and fabricated using standard photo-lithography techniques. In designing microfluidic devices to do what we want, we often have to go through several iterations of the Design → Build → Test cycle. To expedite development, we often use COMSOL Multiphysics to simulate flow characteristics in the device micro-channels. (Read more.)
Drop-Seq: High Throughput Single Cell mRNA Sequencing
We routinely use ‘Drop-Seq’- a high-throughput technique to profile transcriptomes of tens of thousands of single mammalian cells using a combination of DNA barcoding and emulsion microfluidics. This is accomplished by encapsulating and lysing one cell along with a uniquely barcoded microbead per emulsion droplet, that allows molecular tagging of RNA contents from each cell. This technique enables study of the transcription profiles of an unprecedented number of cells at single-cell resolution.
To demonstrate Drop-Seq’s power to categorize cell types in complex tissues, it was applied to mouse retina. ~45,000 single cells from the retina were thus analyzed to computationally assemble an ab initio cell classification of retina cells into 39 sub-types based on their transcription profiles. This classification faithfully reproduced existing molecular, physiological, and anatomical knowledge, while nominating novel classifications for further research. (Read more.)
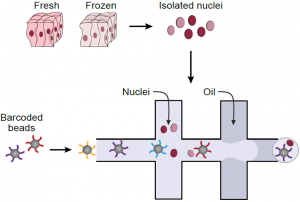 Single nucleus RNA-Seq (sNuc-Seq) profiles RNA in nuclei from tissues that cannot be dissociated or have been previously preserved. Here, we develop DroNc-Seq, a massively parallel sNuc-Seq with droplet technology. DroNc-Seq of 33,284 nuclei from mouse and human archived brain, allowed sensitive, efficient and unbiased recovery of neural and other cell types. DroNc-Seq opens the way for charting systematic cell atlases of complex tissues, including tumor biopsies and human brain. (Read more.)
Single nucleus RNA-Seq (sNuc-Seq) profiles RNA in nuclei from tissues that cannot be dissociated or have been previously preserved. Here, we develop DroNc-Seq, a massively parallel sNuc-Seq with droplet technology. DroNc-Seq of 33,284 nuclei from mouse and human archived brain, allowed sensitive, efficient and unbiased recovery of neural and other cell types. DroNc-Seq opens the way for charting systematic cell atlases of complex tissues, including tumor biopsies and human brain. (Read more.)
Human Cell Atlas: Seed Networks
 The Human Cell Atlas (HCA) project is a recent, world-wide initiative that aims to identify and functionally characterize all cell-types in the human body in health and disease. Single-cell RNA-sequencing (scRNA-seq) methods can be and are being extensively used to profile cells in different tissues and at different developmental stages by their unique gene expression patterns by the HCA. (Read more.)
The Human Cell Atlas (HCA) project is a recent, world-wide initiative that aims to identify and functionally characterize all cell-types in the human body in health and disease. Single-cell RNA-sequencing (scRNA-seq) methods can be and are being extensively used to profile cells in different tissues and at different developmental stages by their unique gene expression patterns by the HCA. (Read more.)
We are grateful to the Chan-Zuckerberg Initiative for their support to the HCA with funds and infrastructure to do so.
A Cell Atlas of Ileal Colonic Crohn’s Disease
 As part of the Human Cell Atlas project, we are working towards identifying and characterizing cell types in the human gut by establishing a comprehensive cellular atlas of the ileum in healthy individuals and patients with ileal-colonic and ileal Crohn’s disease. We are using high-throughput single cell transcriptomic and epigenomic profiling to gut biopsies obtained from patients with CD and healthy controls. These data, integrated with patient metadata and functional characterization of matched organoids, helps generate high-dimensional patient- and disease-specific cellular and transcriptional signatures. This will provide a comprehensive cellular and transcriptomic reference atlas which will allow us to identify cell-type and disease-specific expression patterns and regulatory features. (Read more.)
As part of the Human Cell Atlas project, we are working towards identifying and characterizing cell types in the human gut by establishing a comprehensive cellular atlas of the ileum in healthy individuals and patients with ileal-colonic and ileal Crohn’s disease. We are using high-throughput single cell transcriptomic and epigenomic profiling to gut biopsies obtained from patients with CD and healthy controls. These data, integrated with patient metadata and functional characterization of matched organoids, helps generate high-dimensional patient- and disease-specific cellular and transcriptional signatures. This will provide a comprehensive cellular and transcriptomic reference atlas which will allow us to identify cell-type and disease-specific expression patterns and regulatory features. (Read more.)
We are grateful to the Helmsley Charitable Trust for their support, as part of the Gut Cell Atlas Initiative.
mDrop-Seq: Microbial Single Cell RNA-Seq
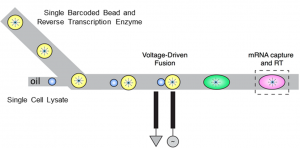
We want to adapt the microfluidic and molecular barcoding techniques used in Drop-Seq to profile single microbial cells in numbers of the order of 105 – 106. We are working on a pico-injection and droplet-merging scheme (see left) to implement microbial Drop-Seq or mDrop-Seq at sufficiently high throughput needed to decipher the heterogeneity in microbial cell populations. (Read more.)
Host-Pathogen Interaction at Controlled MOI
We are developing a novel platform to study the role of cellular heterogeneity on the outcome of host-pathogen interactions using a combination of live-cell imaging and droplet microfluidics.

This will allow us to (1) infect cells with pathogens at a defined Multiplicity Of Infection (MOI) in stable micro-emulsion droplets; (2) incubate and image cells ‘live’ in droplets, where each droplet represents an isolated, stable and reproducible micro-environment; (3) use fluorescent reporters of cell-states to track and sort individual cells, in a “live sorting” assay that can be done repeatedly during infection without disturbing the cell micro-environment; and (4) perform population and single-cell RNA-Seq on both host and pathogen cells. To note, the cells can be easily removed from droplets and re-encapsulated at later time points, thus allowing for long-term studies that allow population interactions like paracrine signaling when desired, but still provide single-cell resolution when needed. The modular compendium of tools developed here will be applicable to diverse host-pathogen systems. (Read more.)
Integrated Silicon Nano-chip/Microfluidics Platform for Microbial ‘Omics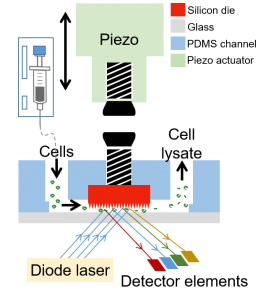
We are working with the Guha lab to integrate semiconductor chip-based components and their functionalities into soft polymeric microfluidics that will enable fast, efficient, high-throughput manipulation of microbial cells and their contents for applications in biotechnology or medicine. The power of chip-integrated photonic, electrical, and micro-mechanical modulation opens a number of capabilities in single cell microbial profiling not available today. We developed a Si Chip-Integrated Soft Microfluidics (SCISM) platform and fabrication technique to perform lysis of microbial cells rapidly and efficiently using micro-mechanical impact without damage to the cell’s fragile contents. (Read more.)
Novel Millifluidic Platform to Study Venous Thrombosis
Vascular access, widely used for hemodialysis and made available through surgically placed arteriovenous fistulas (AVF), are prone to failure because of stenosis and ultimately thrombosis. This contributes to significant morbidity, mortality and cost of care for patients with end-stage renal disease (ESRD). Multiple factors lead to failure of AVF in renal failure including endothelial cell getting activated in the veil wall, high levels of pro-coagulant factors in blood and stagnant blood flow in areas of low wall shear stress.
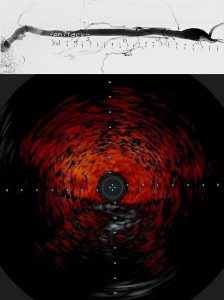
To investigate these factors, we have teamed up with Dr. Mary Hammes, Department of Medicine, Nephrology, to develop a novel millifluidic device and flow platform to make patient-specific vein models that replicate thrombosis. We then use this 3D model to link blood conditions in renal failure using blood from ESRD patients, coagulation factors, and physical and biochemical modeling. Specifically, we are building 3D vein models of the brachio-cephalic arch where thrombosis and stenosis is likely to occur, reconstructed from venogram (top) and intravascular ultrasound (bottom) imaging of the arch in ESRD patients using 3D modelling. The 3D model of the arch is then fabricated into a millifluidic device using 3D printing. (Read more.)
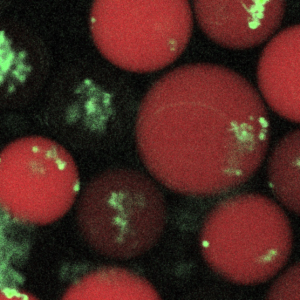
Studies suggest that cellular functions are strongly influenced by their physical surroundings. For example, immune cells like macrophages are substrate dependent in terms of their efficacy in fighting pathogens, while commensals like Candida albicans display different morphology in the presence/absence of key proteins like mucins that are found coated on the epithelia. We want to understand the role of different substrates and their stiffness in either suppressing pathogen directly, or indirectly fighting infection by enhancing the host’s ability to suppress pathogen cells. Interactions between host and pathogenic cells within different biopolymer gels are observed using well-plate assays and droplet microfluidics. The imaging read-out is followed up with whole transcriptome analyses on host-pathogen sub-populations sorted based on infection outcome. (Read more.)
Cell Culture under Gradient Stimuli
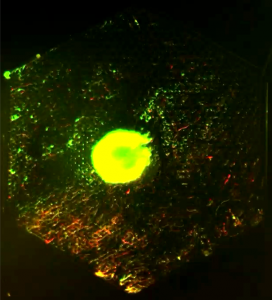
We use PDMS microfluidic device and volume-driven flow to generate a spatial gradient of any chemical of choice on device. This chemical gradient is spatially stable over time, and can be used to maintain a continuous culture of cells under a range of chemical stimuli that can be controlled by adjusting flow-rates. PDMS is gas permeable and bio-compatible that makes it amenable to long-term cell culture (~ weeks). The setup is also compatible with live-cell microscopy and be used to image cell response and growth continuously. To the left is an image of GFP-labeled E. coli that expresses a red fluorescent reporter of nitric oxide that flows in the device in the lower right half of the device. (Read more.)
We use a set of microfluidic devices to test efficacy of cancer chemotherapy drugs on human cancer cell lines where the drug concentrations are systematically varied. Efficacy of drugs is measured as functions of exposure time and drug concentration from the cell fate of cancer cells in each drop, using commercial cell-fate reporter kits. After incubation off chip, we re-inject drops into a second microfluidic chip and use PMT-based fluorophore detection in drops pioneered at Weitz lab to simultaneously read out drug concentration and cell-fate at high throughput. (Read More.)