Crohn’s disease (CD) is a debilitating chronic inflammatory disorder of the gastrointestinal (GI) tract that can present at different locations throughout the entire tract. Involvement of the distal ileum is a common presentation that can progress to complications, including ectopic extensions of mesenteric fat, tissue fibrosis, intestinal obstruction, abscesses, and fistulas. Despite decades of research suggesting that CD arises from a convergence of genetic, environmental, and microbial factors, the cellular and molecular mechanisms leading to CD and the respective contributions of the factors to disease initiation and progression remain poorly understood. The challenge has been that CD are extremely heterogeneous diseases in manifestation as well as response to treatment, and the cellular complexity of the gut make it difficult to identify cellular and molecular drivers of CD.
 As part of the Human Cell Atlas project, we are working towards identifying and characterizing cell types in the human gut by establishing a comprehensive cellular atlas of the ileum in healthy individuals and patients with ileal-colonic and ileal Crohn’s disease. We are using high-throughput single cell transcriptomic and epigenomic profiling to gut biopsies obtained from patients with CD and healthy controls. These data, integrated with patient metadata and functional characterization of matched organoids, helps generate high-dimensional patient- and disease-specific cellular and transcriptional signatures. This will provide a comprehensive cellular and transcriptomic reference atlas which will allow us to identify cell-type and disease-specific expression patterns and regulatory features. This, in turn, will yield insights into the disease etiology, identify key cellular players and might inform novel cellular and molecular disease stratification.
As part of the Human Cell Atlas project, we are working towards identifying and characterizing cell types in the human gut by establishing a comprehensive cellular atlas of the ileum in healthy individuals and patients with ileal-colonic and ileal Crohn’s disease. We are using high-throughput single cell transcriptomic and epigenomic profiling to gut biopsies obtained from patients with CD and healthy controls. These data, integrated with patient metadata and functional characterization of matched organoids, helps generate high-dimensional patient- and disease-specific cellular and transcriptional signatures. This will provide a comprehensive cellular and transcriptomic reference atlas which will allow us to identify cell-type and disease-specific expression patterns and regulatory features. This, in turn, will yield insights into the disease etiology, identify key cellular players and might inform novel cellular and molecular disease stratification.
We are a multi-disciplinary team with diverse, complementary expertise covering clinical and basic research of Inflammatory Bowel Diseases (Chang Lab), single-cell epigenomics and transcriptomics- us, and Dr. Sebastian Pott, and computational biology and statistics (Dr. Matthew Stephens’ Lab). This project also takes advantage of the Inflammatory Bowel Disease clinical pipeline, with cores for patient recruitment and acquisition of surgical and endoscopic tissue samples by the combined GI and colo-rectal surgery group headed by Dr. David Rubin at the University of Chicago.
A comprehensive cellular atlas of the human gut must reflect the cellular composition in vivo and account for variability between individuals and differences in sampling locations. We are creating a detailed molecular and functional atlas of cells in the terminal ileum and proximal colon using samples from individuals with and without CD. We restrict ourselves to these regions where the largest subset of CD patients develops disease.
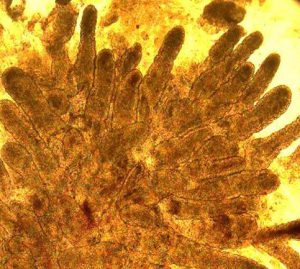
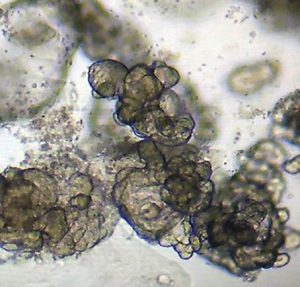
While primary tissues like colonoscopic biopsies (above, left) obtained fresh from the clinic, are invaluable for insights into cellular composition and transcriptional and regulatory states present in vivo, they are fragile and limited in quantity for studying patient-specific responses to different disease-relevant stimuli. To bypass this limitation, we are establishing patient-derived organoids (above, right; courtesy Dr. Candace Cham) as a powerful model to study transcriptional response to different stimuli to serve as patient avatars for individualized medicine approaches.
We aim to
- Identify cell-types in ileum and proximal colon in individuals with and without CD,
- Quantify changes in the cellular composition,
- Measure cell-type specific changes in gene expression,
- Relate transcription to regulatory states,
- Use cell-type and disease-specific expression patterns and chromatin accessibility to annotate GWAS-identified variants,
- Identify cell-types and cell-type specific expression and regulatory features associated with fibrotic tissues and creeping fat in CD,
- Identify mechanisms that modulate transcriptional response in patient-derived organoids.
Some stories featuring this project and other GCA initiatives are found here, here, and here.
 We are grateful to the Helmsley Charitable Trust for their support, as part of the Gut Cell Atlas Initiative.
We are grateful to the Helmsley Charitable Trust for their support, as part of the Gut Cell Atlas Initiative.