Single cell genomics is a rapidly emerging field with extensive use in understanding fundamental biology and applications in medicine, and industry. However, these technologies have largely failed to translate to the world of microbes that include fungi, bacteria, etc. and the microbiome they constitute. Small cell size and strong cell walls make it difficult to lyse individual microbial cells needed for single cell genomics, proteomics or metabolomics (broadly described as ’omics) applications. Currently, microbial cell lysis needs to be tailored to particular microbial species or strains and uses chemical cocktails that are slow to act; there are no rapid, high throughput single-cell RNA sequencing (scRNA-seq) techniques that can be applied to multiple microbial species in an unbiased way. Such assays of the different microbial species and strains in a single shot and at high-throughput can reveal the constituent players as well as their genomic and biochemical activity at the time of sampling. Potential applications are far-ranging: from understanding the functions of the human microbiome in health and disease, plant and soil microbiome in agriculture, to characterizing previously unknown ecological niches and production of biologics.
To address this challenge and opportunity, we are working with the Guha lab to integrate semiconductor chip-based components and their functionalities into soft polymeric microfluidics that will enable fast, efficient, high-throughput manipulation of microbial cells and their contents for applications in biotechnology or medicine. The power of chip-integrated photonic, electrical, and micro-mechanical modulation opens a number of capabilities in single cell microbial profiling not available today.
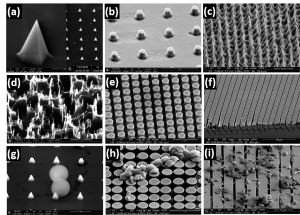
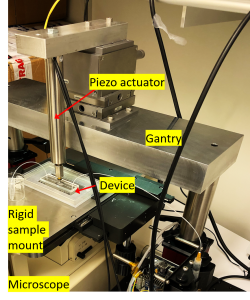 We developed a Si Chip-Integrated Soft Microfluidics (SCISM) platform and fabrication technique to perform lysis of microbial cells rapidly and efficiently using micro-mechanical impact without damage to the cell’s fragile contents. To accomplish this, a silicon chip, containing an array of geometric features is embedded in a microfluidic device and driven vertically back and forth by a piezoelectric transducer at high frequency. This causes the array to press upon and perforate or crack the cell wall of microbes flowing through the fluidic channel directly beneath the chip. The microfluidic device is fabricated using a soft elastomer that is cast from a 3D-printed mold. Geometric features on our silicon chips incorporated into our hybrid devices include- see figure below- sharp cones (a), truncated cones (b), cryo-etched needles (c), corrugated black silicon (d), cylinders (e), and ridges (f). We then tested these designs for their relative efficiency in rupturing polymer or porous silica microbeads of different size and hardness (g-i), as proxy for microbes. A syringe pump is used to drive flow of microbeads/microbes in the microfluidic channel; controller unit drives the Si chip independently using piezoelectric actuation. The setup is coupled with an optical microscope to visualize the piezo-driven mechanical impaction on the microbes/microbeads in the device.
We developed a Si Chip-Integrated Soft Microfluidics (SCISM) platform and fabrication technique to perform lysis of microbial cells rapidly and efficiently using micro-mechanical impact without damage to the cell’s fragile contents. To accomplish this, a silicon chip, containing an array of geometric features is embedded in a microfluidic device and driven vertically back and forth by a piezoelectric transducer at high frequency. This causes the array to press upon and perforate or crack the cell wall of microbes flowing through the fluidic channel directly beneath the chip. The microfluidic device is fabricated using a soft elastomer that is cast from a 3D-printed mold. Geometric features on our silicon chips incorporated into our hybrid devices include- see figure below- sharp cones (a), truncated cones (b), cryo-etched needles (c), corrugated black silicon (d), cylinders (e), and ridges (f). We then tested these designs for their relative efficiency in rupturing polymer or porous silica microbeads of different size and hardness (g-i), as proxy for microbes. A syringe pump is used to drive flow of microbeads/microbes in the microfluidic channel; controller unit drives the Si chip independently using piezoelectric actuation. The setup is coupled with an optical microscope to visualize the piezo-driven mechanical impaction on the microbes/microbeads in the device.
Overall performance of the SCISM platform is optimized by simultaneously adjusting the frequency and amplitude of the piezoelectric transducer, pump flow rates, micro-bead (or microbe density), etc. We used this stetup to perform lysis of Saccharomyces cerevisiae and Candida albicans cells, a model yeast microbe, using qRT-PCR. See the pre-print here.
Future manifestation of the SCISM platform can contain electrical, photonic and/or mechanical probes for accessing single cells at sub-micron or nano-scale. These elements will be modular in design and may be used for various purposes, such as measuring physical properties, biochemical contents, or fluidic manipulation of the microbes. As next step, we are integrating on-chip Raman spectroscopy to classify the cells’ transcriptional states via the RNA’s vibrational modes.