The Human Cell Atlas (HCA) project is a recent, world-wide initiative that aims to identify and functionally characterize all cell-types in the human body in health and disease. High throughput state-of-the-art sequencing and imaging techniques like single-cell RNA sequencing (scRNA-seq), Assay for single-cell Transposase-Accessible Chromatin (scATAC-seq) and spatial transcriptomics methods can be and are being extensively used to profile cells in different tissues and at different developmental stages by their unique gene expression patterns by the HCA. Seed networks spanning multiple institutions across multiple countries have formed to take on this challenge, each focused on a specific organ of interest.
The Human Cell Atlas (HCA) project is a recent, world-wide initiative that aims to identify and functionally characterize all cell-types in the human body in health and disease. High throughput state-of-the-art sequencing and imaging techniques like single-cell RNA sequencing (scRNA-seq), Assay for single-cell Transposase-Accessible Chromatin (scATAC-seq) and spatial transcriptomics methods can be and are being extensively used to profile cells in different tissues and at different developmental stages by their unique gene expression patterns by the HCA. Seed networks spanning multiple institutions across multiple countries have formed to take on this challenge, each focused on a specific organ of interest.
Based on different interests of our lab members and using the single-cell techniques benchmarked by us and others in the CZI pilot experiments, we are working on three organs of interest, viz., the heart, female reproductive system and gut. As part of the CZI Seed Networks, we are proud to participate in the respective Seed Networks to contribute to and augment our efforts.
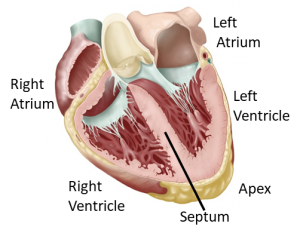 Heart: Along with a multi-national team of cardiologists, technology developers and bioinformatics experts led by Dr. Norbert Hubner from Max Delbruck Center in Berlin, Germany and in close collaboration with Dr. Sebastian Pott, we are creating a detailed cellular and molecular map of 6 distinct anatomical regions of the human heart (as marked in the image to the right) using adult post mortem tissue. B
Heart: Along with a multi-national team of cardiologists, technology developers and bioinformatics experts led by Dr. Norbert Hubner from Max Delbruck Center in Berlin, Germany and in close collaboration with Dr. Sebastian Pott, we are creating a detailed cellular and molecular map of 6 distinct anatomical regions of the human heart (as marked in the image to the right) using adult post mortem tissue. B ecause of challenges unique to adult cardiomyocytes, including cell size (100-150 microns along the long axis), shape (rod-like) and difficult lysis, usual single cell RNA sequencing techniques do not work on these cells. Since the majority of the cells in our regions of interest are composed of cardiomyocytes, we are using single nuclei harvested from the tissues and DroNc-seq to generate this anatomical map.
ecause of challenges unique to adult cardiomyocytes, including cell size (100-150 microns along the long axis), shape (rod-like) and difficult lysis, usual single cell RNA sequencing techniques do not work on these cells. Since the majority of the cells in our regions of interest are composed of cardiomyocytes, we are using single nuclei harvested from the tissues and DroNc-seq to generate this anatomical map.
Interestingly, cardiomyocytes are also atypical in that they are often multi-nucleated, but it is not clear if the different nuclei from a given cell perform the same functions or specializes in certain tasks. Concurrently, we are developing a customized Drop-seq protocol that can tackle the large cell-size and difficult lysis challenges and enable ‘single cell’ RNA-seq on adult cardiomyocytes. Together with single nucleus RNA-seq data on the same cardiomyocyte cells, we hope to learn the role of multiple nucleus in these fascinating cells.
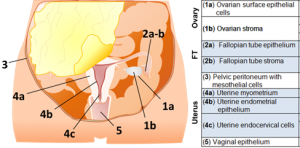
Female Reproductive System: Together with Dr. Ernst Lengyel, a world leader in Ovarian Cancer, and Dr. Mengjie Chen, we are creating a cell atlas of the female reproductive system, with focus on ovary, Fallopian tube, uterus, peritoneum and vagina. We are interested in studying the stromal and epithelial compartments and posit that this study will establish a healthy baseline for future studies on gynecological diseases including our own ovarian cancer studies.
Gut: As part of the Helmsley’s Gut Cell Atlas initiative, we are generating a detailed cellular and functional map of the ileum and its adjoining ascending colon. Read more.
We are grateful to the Chan-Zuckerberg Initiative and the Helmsley Charitable Trust for their support to the HCA with funds and infrastructure to do so.